2025
Learned K-Space Partitioning for Improved Dual-Domain Self-Supervised Image Reconstruction
Kadota B., Millard C., Chiew M.
Conference in IEEE Engineering in Medicine and Biology (in press)
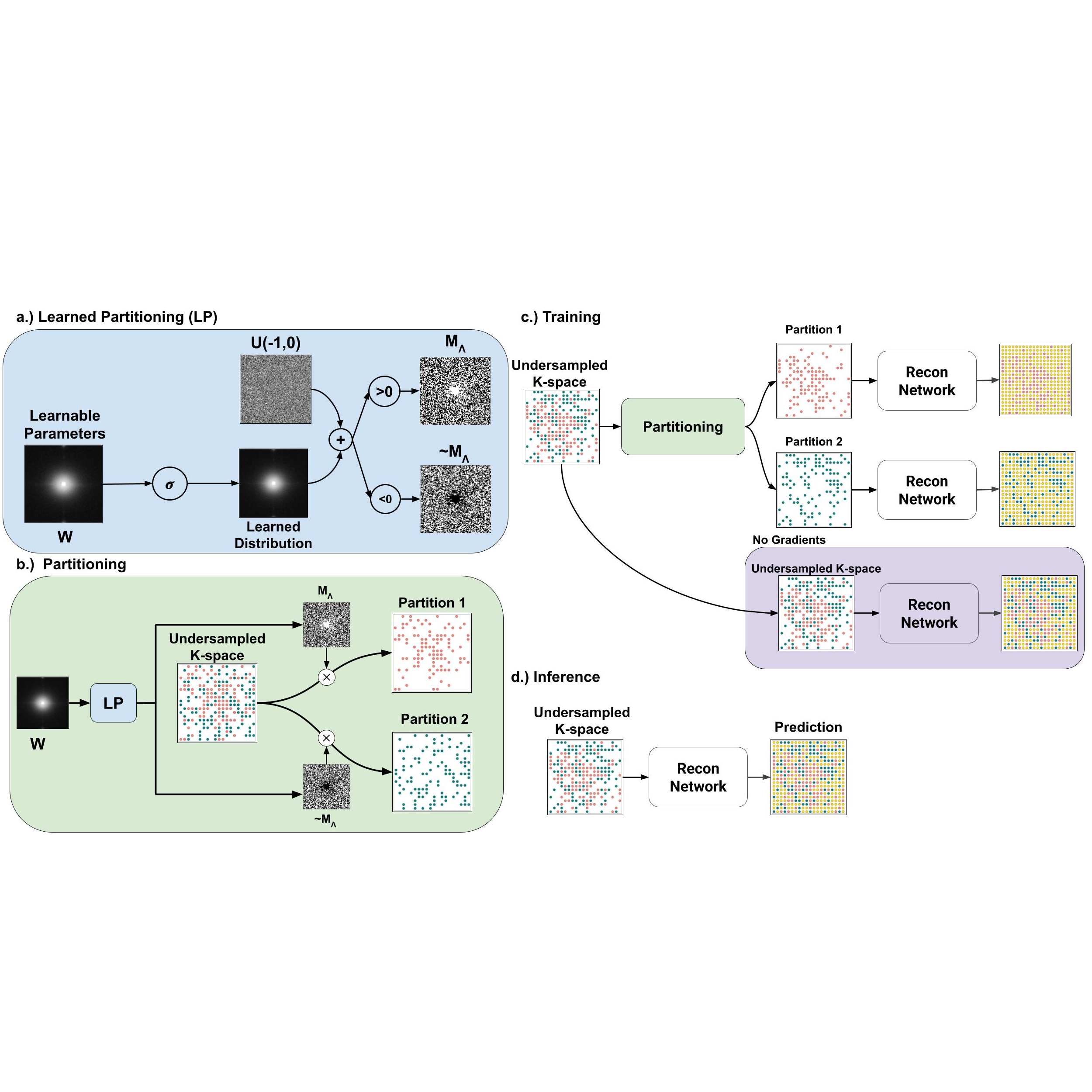
Self-supervised magnetic resonance imaging (MRI) reconstruction methods can train deep learning networks without requiring fully sampled reference data. One such approach, self-supervised via data under-sampling (SSDU), partitions under-sampled k-space into two disjoint sets, with a neural network mapping between them. However, SSDU and its variants rely on heuristic k-space partitioning, which may lead to suboptimal performance and necessitates new partitioning schemes when initial under-sampling patterns change. In this work, we propose a novel approach to learn optimal k-space partitioning by modeling a probability distribution which we use for partitioning. Specifically, we employ the LOUPE framework to learn an optimal partitioning probability distribution. Furthermore, we introduce a weighted dual-domain self- supervised loss function that incorporates both k-space and image-space loss terms. Evaluations on the fastMRI dataset demonstrate that our dual-domain learned partitioning method outperforms existing partitioning strategies and adapts to new sampling patterns without requiring hand-picked partitioning methods.
2024
Low-Rank Tensor U-Net for Accelerated Cardiac MR Imaging
Patel J., Kadota B., Sheagren C., Chiew M., Wright G.
International Workshop on Statistical Atlases and Computational Models of the Heart
Cardiovascular diseases (CVDs) remain the leading cause of mortality and morbidity worldwide. Both diagnosis and prognosis of these diseases benefit from high-quality imaging, which cardiac magnetic resonance imaging provides. CMR imaging requires lengthy acquisition times and multiple breath-holds for a complete exam, which can lead to patient discomfort and frequently results in image artifacts. In this work, we present a Low-rank tensor U-Net method (LowRank-CGNet) that rapidly reconstructs highly undersampled data with a variety of anatomy, contrast, and undersampling artifacts. The model uses conjugate gradient data consistency to solve for the spatial and temporal bases and employs a U-Net to further regularize the basis vectors. Currently, model performance is superior to a standard U-Net, but inferior to conventional compressed sensing methods. In the future, we aim to further improve model performance by increasing the U-Net size, extending the training duration, and dynamically updating the tensor rank for different anatomies.
2023
Retrospective frequency drift correction of rosette MRSI data using spectral registration
Kadota B, Millard C, Chiew M
Magnetic Resonance in Medicine
Purpose: Frequency drift correction is an important postprocessing step in MRS that yields improvements in spectral quality and metabolite quantification. Although routinely applied in single-voxel MRS, drift correction is much more challenging in MRSI due to the presence of phase-encoding gradients. Thus, separately acquired navigator scans are normally required for drift estimation. In this work, we demonstrate the use of self-navigating rosette MRSI trajectories combined with time-domain spectral registration to enable retrospective frequency drift corrections without the need for separately acquired navigator echoes.
Methods: A rosette MRSI sequence was implemented to acquire data from the brains of 5 healthy volunteers. FIDs from the center of k-space (k = 0 FIDs) were isolated from each shot of the rosette acquisition, and time-domain spectral registration was used to estimate the frequency offset of each k = 0 FID relative to a reference scan (the first k = 0 FID in the series). The estimated frequency offsets were then used to apply corrections throughout k -space. Improvements in spectral quality were assessed before and after drift correction.
Results: Spectral registration resulted in significant improvements in signal-to-noise ratio (12.9%) and spectral linewidths (18.5%). Metabolite quantification was performed using LCModel, and the average Cramer-Rao lower bounds uncertainty estimates were reduced by 5.0% for all metabolites, following field drift correction.
Conclusion: This study demonstrated the use of self-navigating rosette MRSI trajectories to retrospectively correct frequency drift errors in in vivo MRSI data. This correction yields meaningful improvements in spectral quality.
2023
Accelerated Cardiac Parametric Mapping Using Deep Learning-Refined Subspace Models
Sheagren C., Kadota B., Patel J., Chiew M., Wright., G
International Workshop on Statistical Atlases and Computational Models of the Heart
Cardiac parametric mapping is useful for evaluating cardiac fibrosis and edema. Parametric mapping relies on single-shot heartbeat-by-heartbeat imaging, which is susceptible to intra-shot motion during the imaging window. However, reducing the imaging window requires undersampled reconstruction techniques to preserve image fidelity and spatial resolution. The proposed approach is based on a low-rank tensor model of the multi-dimensional data, which jointly estimates spatial basis images and temporal basis time-courses from an auxiliary parallel imaging reconstruction. The tensor-estimated spatial basis is then further refined using a deep neural network, trained in a fully supervised fashion, improving the fidelity of the spatial basis using learned representations of cardiac basis functions. This two-stage spatial basis estimation will be compared against Fourier-based reconstructions and parallel imaging alone to demonstrate the sharpening and denoising properties of the deep learning-based subspace analysis.
2023
Jérôme Waldispühl, When online citizen science meets teaching: Storyfication of a science discovery game to teach, learn, and contribute to genomic research
C. Drogaris, A. Butyaev, E. Nazarova, R. Sarrazin-Gendron, H. Patel, A. Singh, B. Kadota.
Biochemistry and Molecular Biology Education,
In the last decade, video games became a common vehicle for citizen science initiatives in life science, allowing participants to contribute to real scientific data analysis while learning about it. Since 2010, our scientific discovery game (SDG) Phylo enlists participants in comparative genomic data analysis. It is frequently used as a learning tool, but the activities were difficult to aggregate to build a coherent teaching activity. Here, we describe a strategy and series of recipes to facilitate the integration of SDGs in courses and implement this approach in Phylo. We developed new roles and functionalities enabling instructors to create assignments and monitor the progress of students. A story mode progressively introduces comparative genomics concepts, allowing users to learn and contribute to the analysis of real genomic sequences. Preliminary results from a user study suggest this framework may help to boost user motivation and clarify pedagogical objectives.